Argumentation System and User Guide
About the tool
A. Groza, O. Popa - Mining Arguments from
Cancer Documents Using Natural Language Processing and
Ontologies. IEEE 12
International Conference on Intelligent Computer
Communication and Processing (ICCP2016), Cluj-Napoca, Romania,
8-10 September 2016.(pdf)
In the medical domain, the continuous stream of scientific research contains contradictory results supported by arguments and counter-arguments. As medical expertise occurs at different levels, part of the human agents have difficulties to face the huge amount of studies, but also to understand the reasons and pieces of evidences claimed by the proponents and the opponents of the debated topic. To better understand the supporting arguments for new findings related to current state in the medical domain we need tools able to identify arguments in scientific papers. The system relies on the well-known domain knowledge interleaved with the natural language processing. To formalise the existing medical knowledge, the system relies on ontologies. To structure the argumentation model the system uses also the expressivity and reasoning capabilities of Description Logics. To perform argumentation mining the system uses various linguistic patterns in a rule-based language. The solution was tested against a corpus of scientific papers related to breast cancer.
The main feature of the tool is to detect automatically the structure of the arguments inside medical documents. An argument is formed by one claim and one or more premises, this instrument is capable of detecting this sentences or parts using an algorithm based on the presence of the coordinating conjunction.
In the medical domain, the continuous stream of scientific research contains contradictory results supported by arguments and counter-arguments. As medical expertise occurs at different levels, part of the human agents have difficulties to face the huge amount of studies, but also to understand the reasons and pieces of evidences claimed by the proponents and the opponents of the debated topic. To better understand the supporting arguments for new findings related to current state in the medical domain we need tools able to identify arguments in scientific papers. The system relies on the well-known domain knowledge interleaved with the natural language processing. To formalise the existing medical knowledge, the system relies on ontologies. To structure the argumentation model the system uses also the expressivity and reasoning capabilities of Description Logics. To perform argumentation mining the system uses various linguistic patterns in a rule-based language. The solution was tested against a corpus of scientific papers related to breast cancer.
The main feature of the tool is to detect automatically the structure of the arguments inside medical documents. An argument is formed by one claim and one or more premises, this instrument is capable of detecting this sentences or parts using an algorithm based on the presence of the coordinating conjunction.
Installation Guide
In this section the main installation and
configuration issues for this system are addresed. For
using the tool, the user needs to have installed on the PC
the following tools:
- Eclipse IDE
- Racer reasoner
- Protege 5.0.0
- GATE Developer 8.0
- Check if the computer has a version of JDK 7 or higher;
- Check if the machine has a GATE framework installed. If it doesn't exists this can be download from here;
- Add GATE_HOME as environment variable, by setting it to the folder location where GATE is installed, see Figure 1;
- Create a new folder where the source code is copied to the system;
- Check if the machine has installed the RacerPro application. If it doesn't exists this can be download from here;
- Check if the machine has installed the Protege application. If it doesn't exists this can be download from here;
- Download an Eclipse IDE;
- Import the project in Eclipse with the File -> Import option;
- In the window Package Explorer right click on the name of the project and then select the option Build Path -> Configure Build Path, see Figure 2;
- As seen in the Figure 3, add the JARS existed in the lib folder of the project;
- Select the option Project -> Build Project.
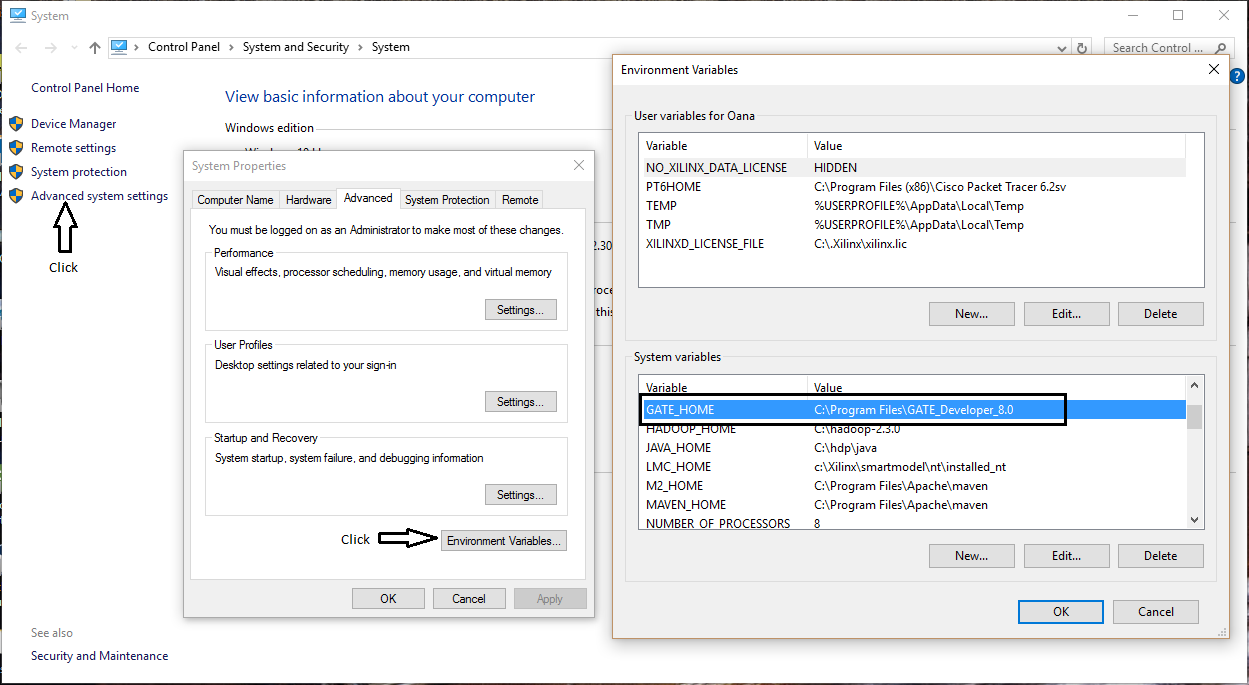
Figure 1. Adding the
environment variable GATE_HOME;
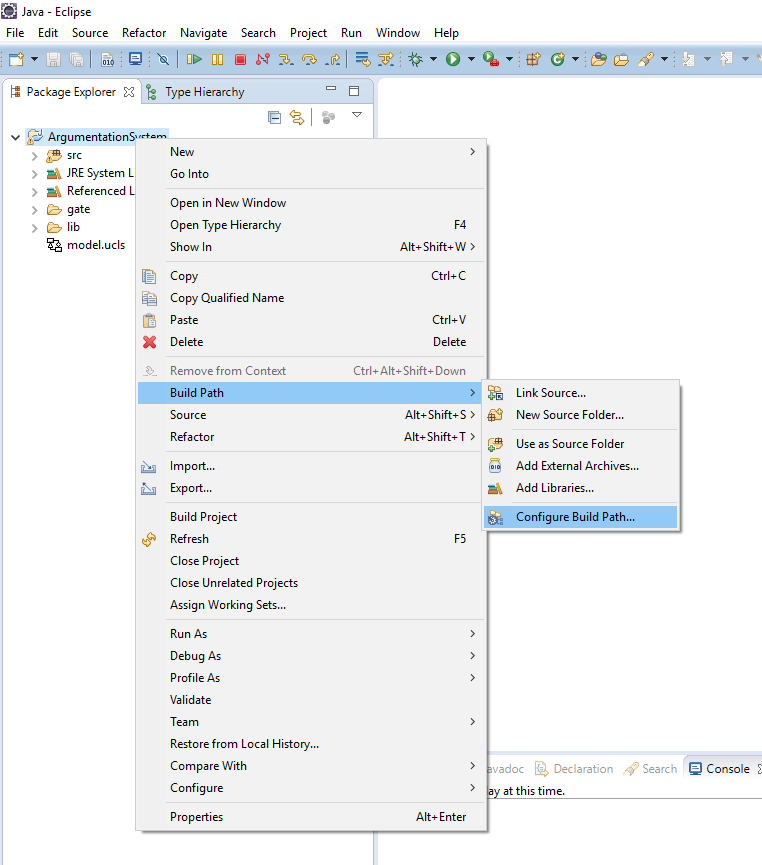
Figure 2. Update the build
path
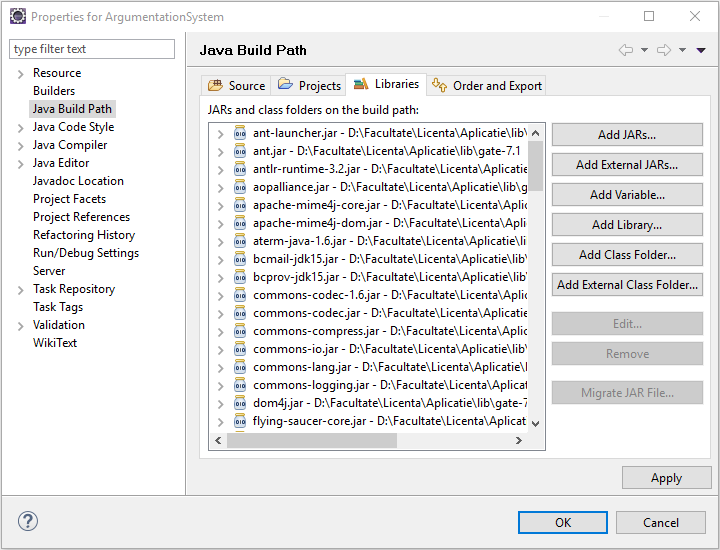
Figure 3. Adding or removing
libraries;
- A. Groza, O. Popa - Mining Arguments from Cancer Documents Using Natural Language Processing and Ontologies, ICCP2016, 8-10 September 2016, Cluj-Napoca (pdf)
- Oana Maria Popa, DETECTAREA
ARGUMENTELOR DIN CADRUL DOCUMENTELOR DESPRE CANCER
UTILIZAND PROCESAREA LIMBAJULUI NATURAL SI ONTOLOGII
, Bachelor thesis, Computer Science Department,
Technical University of Cluj-Napoca, 2016
User's manual
This section describes the main
functionality of the Argumentation System.
Adrian.Groza [at] cs.utcluj.ro
Running the system
This action will be taken
just to the first run of the project. Steps:
- Open the project with Eclipse IDE;
- Change the path for the GATE and Protege applications with the location on the current system;
- Copy from the gazetteer and NE folders found at Aplicatie -> gate -> grammar to destination folders, in this case at C: -> Program Files -> GATE_Developer_8.0 -> plugins -> ANNIE -> resources -> gazetteer and C: -> Program Files -> GATE_Developer_8.0 -> plugins -> ANNIE -> resources -> NE;
- Run the application using the Run option from the Run menu;
Arguments
identification
The output of this action
will be the new entries in the argumentation
ontology.
- Select from the Racer menu the option Start RacerPorter, this command starting the application RacerPro;
- Select from the Cancer ontology menu the option Load ontology. On the screen will appear a File Chooser. As seen in Figure 4, navigate to the location of the breast cancer ontology (Aplicatie -> gate -> ontologies -> breastCancer.racer) and select it;
- Click on the Open button, after which a message will be visible as seen in Figure 5;
- Select the option Create lists related to cancer from the Cancer ontology menu;
- Select the option Close connection ontology from the Cancer ontology menu;
- Select from the Arguments menu the option Load ontology. On the screen will appear a File Chooser;. As seen in Figure 6, navigate to the location of the breast cancer ontology (Aplicatie -> gate -> ontologies -> ontologieArgumente.racer) and select it;
- Click on the Open button, after which a message will be visible as seen in Figure 7;
- Select the option Create lists related to arguments from the Arguments ontology menu;
- Select the option Set corpus from the Arguments ontology menu, this being used to set up the corpus of documents inside the GATE application;
- Select the option Identify claim and Identify premise from the Arguments ontology menu;
- Select the option Save ontology from the Arguments ontology menu to save the new arguments detected;
- Select the option Close connection ontology from the Arguments ontology menu;
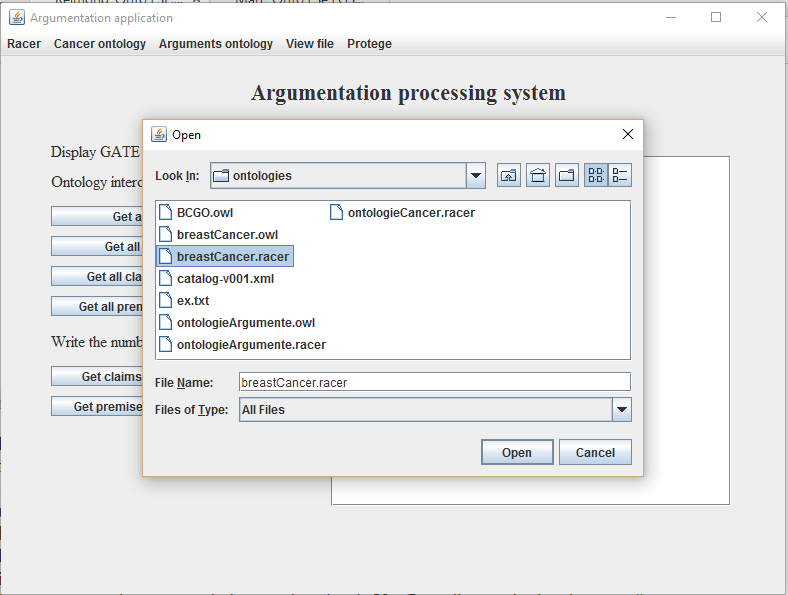
Figure 4. Adding cancer
ontology
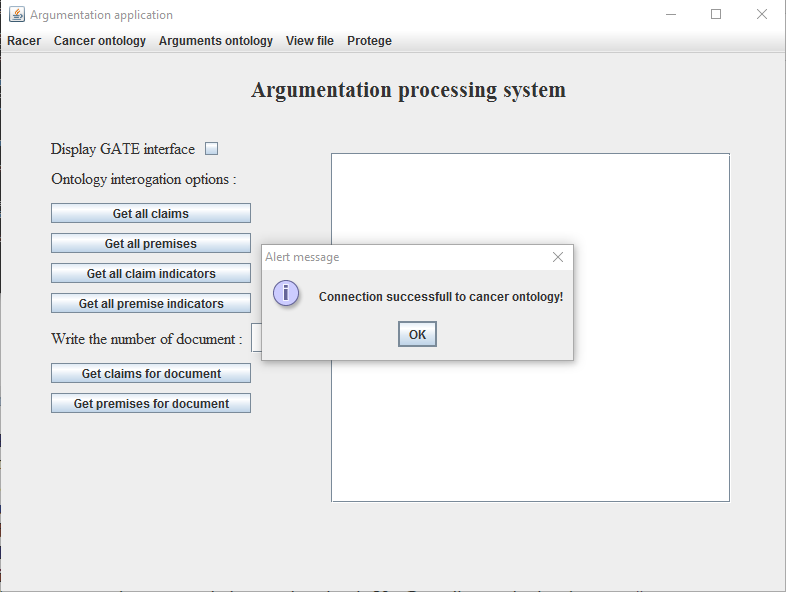
Figure 5. Successfully
uploaded breast cancer ontology
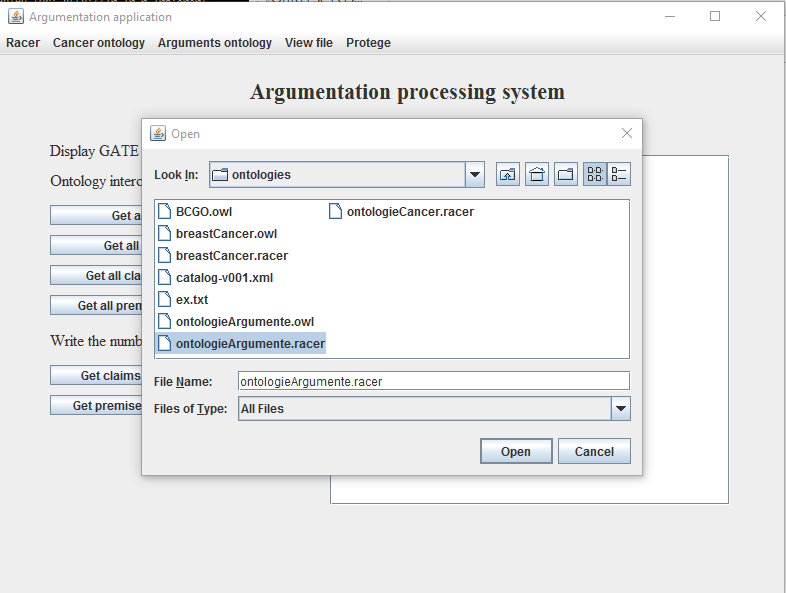
Figure 6. Adding
argumentation ontology
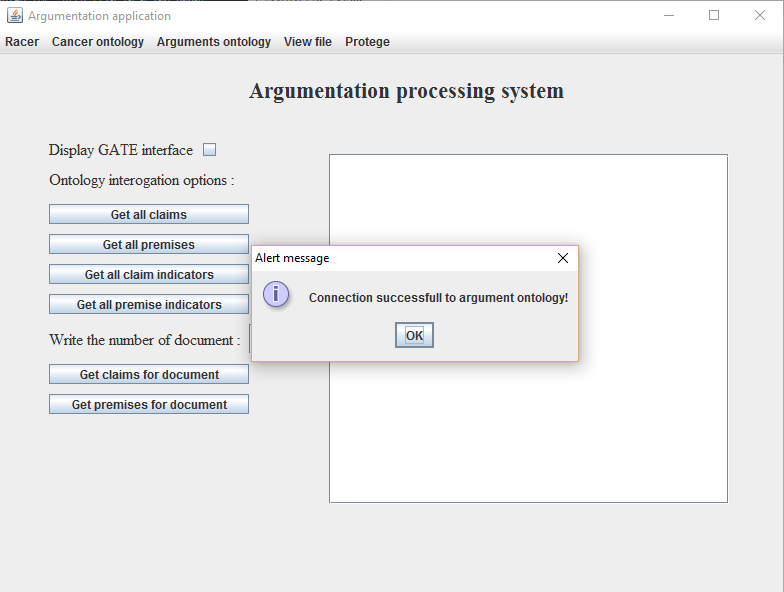
Figure 7. Successfully
uploaded argumenation ontology
Querying the arguments
ontology
In this action the user can
interogate the newly arguments identified using
buttons, the result being displayed on the screen.
- As seen in Figure 8, to display all the claim press on the button called Get all claims present on the graphical interface;
- To detect claims or premises from a specific document write a number between 0 and 5 inside the empty box behind the text Write the number of the document :. If an error occured a message will appear, as seen in Figure 9;
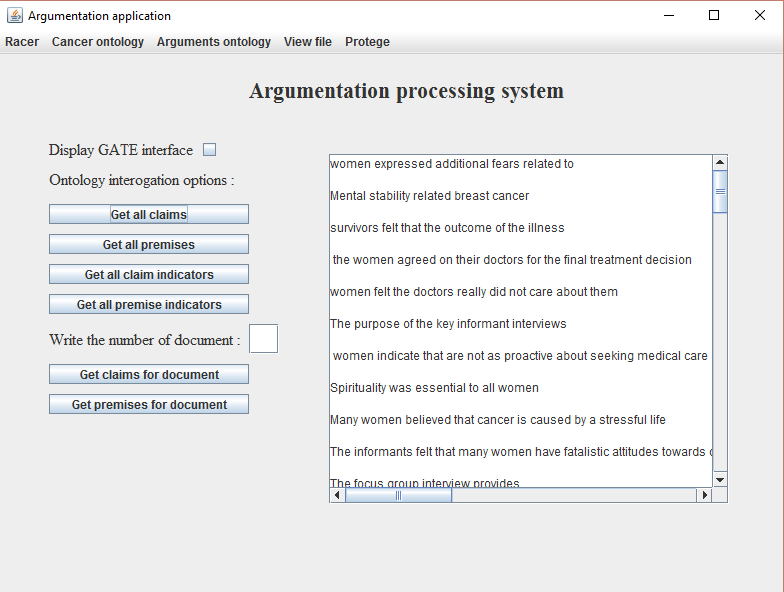
Figure 8. Display all
the claims from the corpus
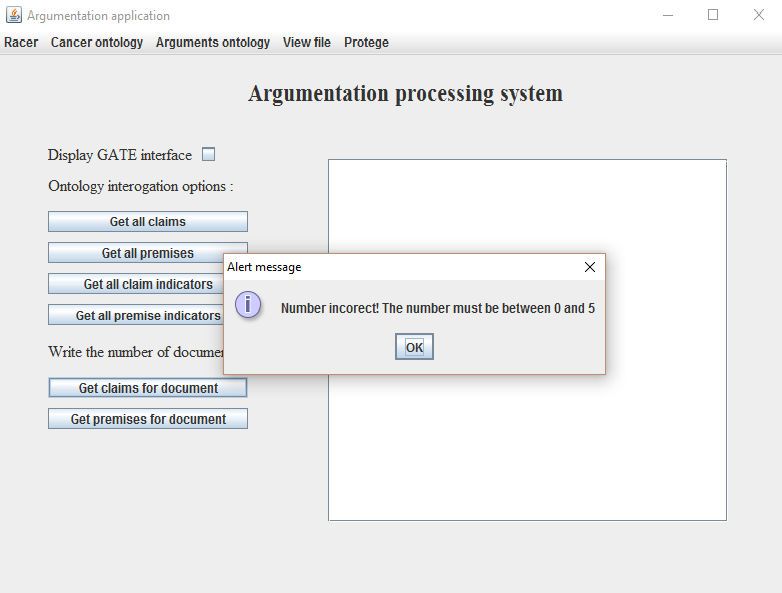
Figure 9. Error message
displayed on the wrong writting of a character
Visualising the
adnotation with GATE
With this action the
adnotation can be seen inside the text documents
undelined with different colors.
- Check the checkbox located behind the text Display GATE interface;
- Select a document from the left side, named Language Resources after the interface of the GATE application will appear;
- Press the Application Sets tab and select different annotation to visualize them, as seen in Figure 10;
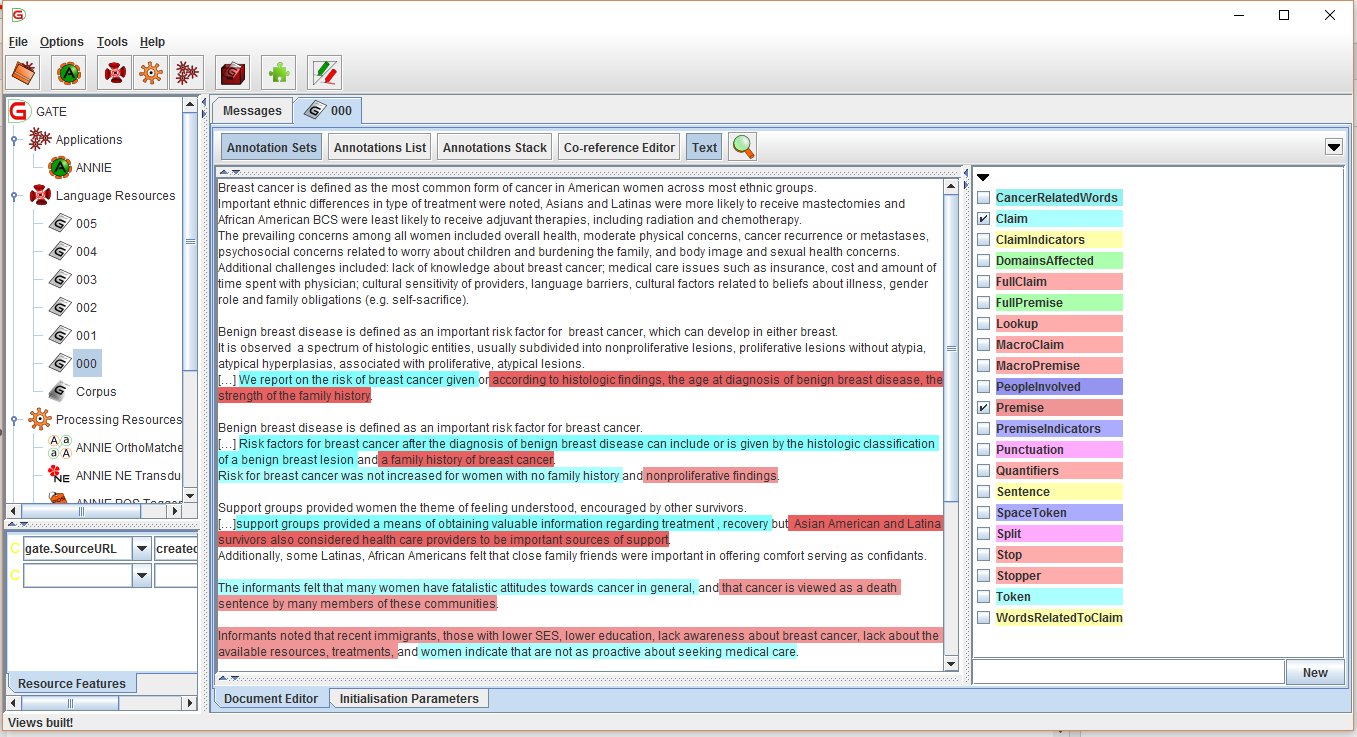
Figure 10. Visualising
the adnotations
Visualising the
ontologies
The ontologies can be seen
with the help of Protege application like graphs;
but this can be done just after the transformation
from the .racer to .owl extension.
- Select the option Convert cancer ontology to owl from the Protege menu;
- Select the option Convert arguments ontology to owl from the Protege menu;
- Select Start Protege from the Protege menu;
- Choose one of the files resulted above to load inside this tool using the options File -> Open;
- From the menu Window select Views -> Ontology views -> OntoGraf import View;
- To visualize a specific entity double click on it, as seen in the Figure 11;
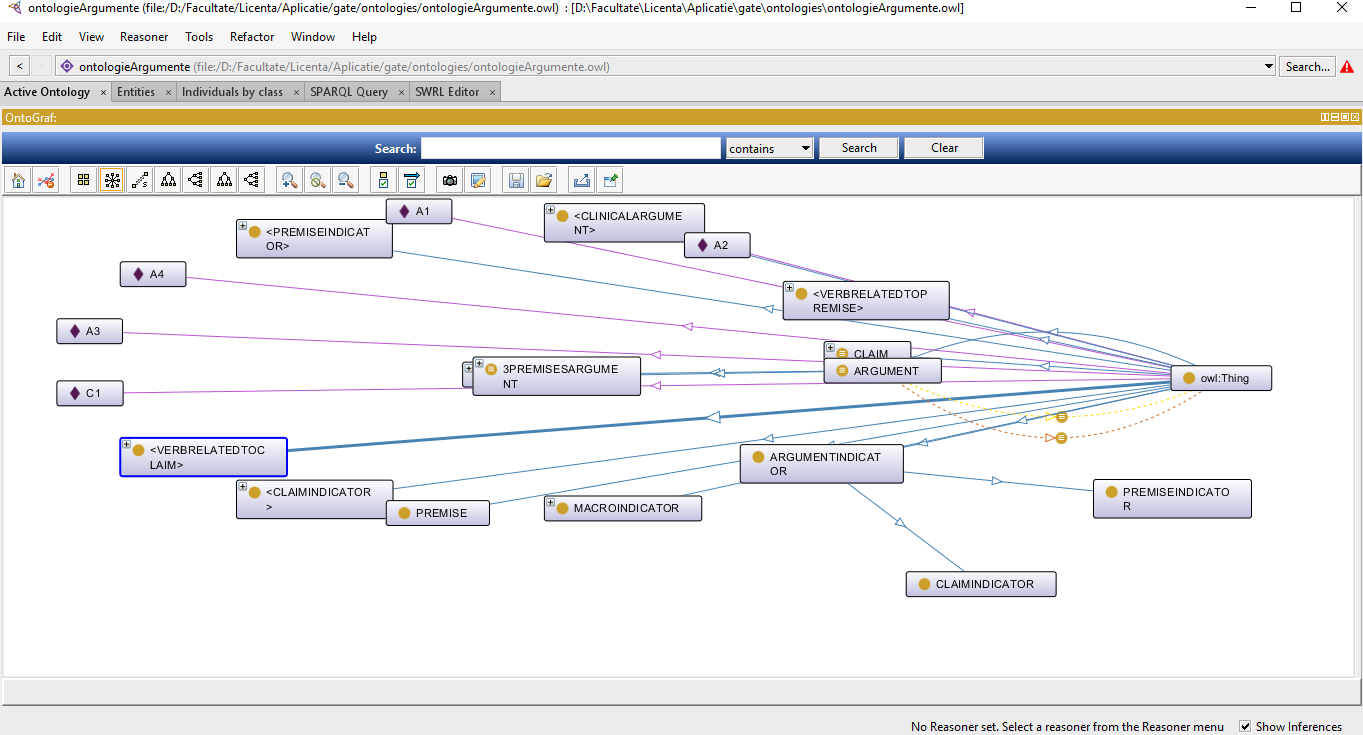
Figure 11. Visualising
the argumenation ontology using Protege
application
Visualising the lists
of words
For visualising the lists of
words, .lst follow the next steps:
- Select from the menu View file the option Open file. On the screen will appear a File chooser by which can navigate to the location of the files, in this case being C: -> Program Files -> GATE_Developer_8 -> plugins > ANNIE -> resources -> gazetteer;
- Select a list of words and then click to the button Open;
- Following this steps on the screen will appear an image like in the Figure 12;
- To delete the content from the box choose from the menu View file the option Clear text field;
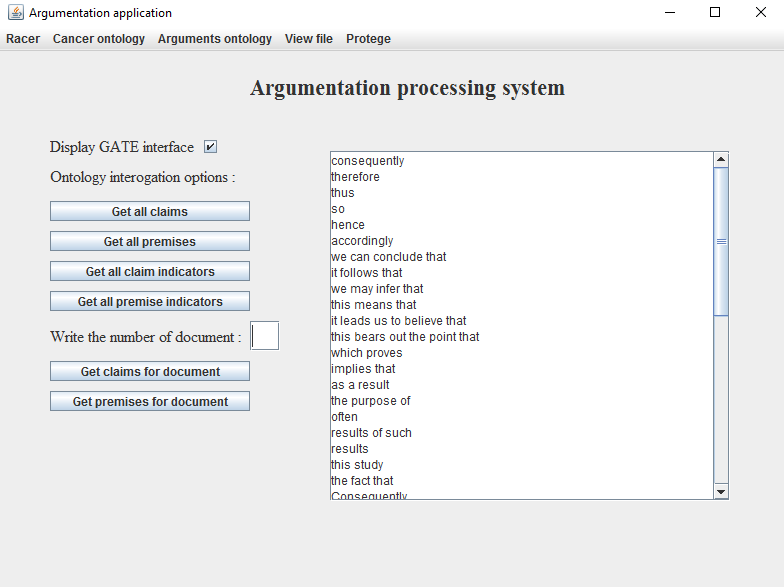
Figure 12. Visualising
the lists of words
Contact
oana_pdv [at] yahoo.com Adrian.Groza [at] cs.utcluj.ro